BIOCHEMICAL CHARACTERIZATION
| main |
| behavior |
| pheromone |
| receptors |
| neural response |
| publications |
| people |
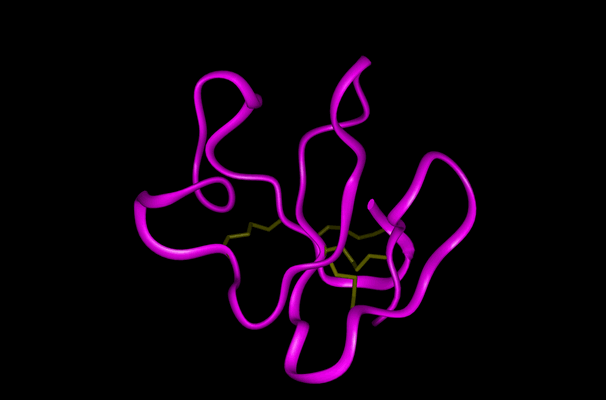 |
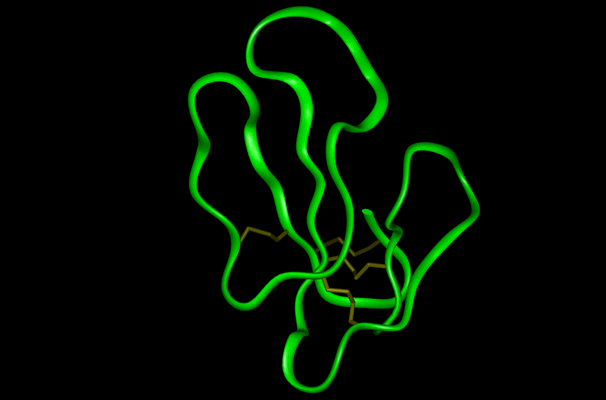
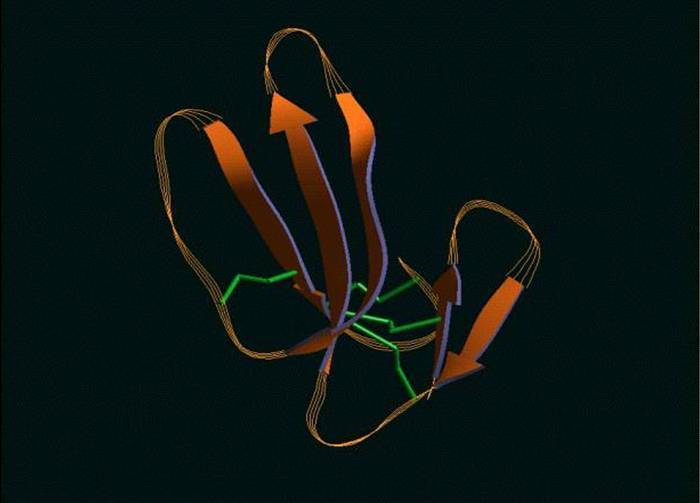
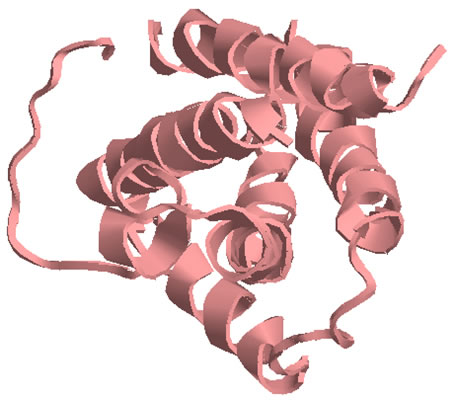
A theoretical structural model of PMF-H. The Insight II homology modeling program, Modeler (Accelrys Software, Inc., San Diego, CA) was used to construct the PMF structural model based on the known crystal structure of cardiotoxin V chain A as determined by Sun et al. (ID# 1KXI in the Protein Data Bank at http://www.rcsb.org). Cardiotoxin V from the Taiwan cobra (Naja naja atra) is a member of the snake venom cytotoxin functional class of small (~7 kDa), highly disulfide-bonded proteins that possess a three-finger structural motif and a 1-3, 2-4, 5-6, 7-8 disulfide bonding pattern.
|



| Characterization of the Major Pheromone Components |
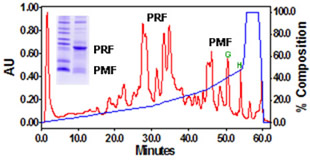
BIOCHEMICAL AND PROTEOMIC STUDIES Biochemical investigations of the glandular extract demonstrated that the pheromone mixture consists of two major families, each of which contains multiple isoforms (Fig. 1). The 22 kDa family (Plethodontid Receptivity Factor; PRF) is related to the IL-6 family of cytokines, while the 7 kDa family (Plethodontid Modulating Factor; PMF) is related to xenoxins and snake toxins.
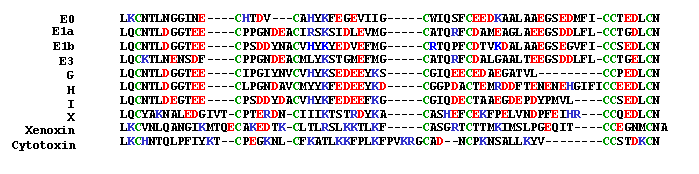
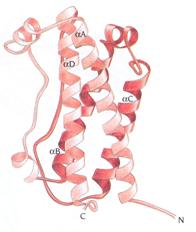
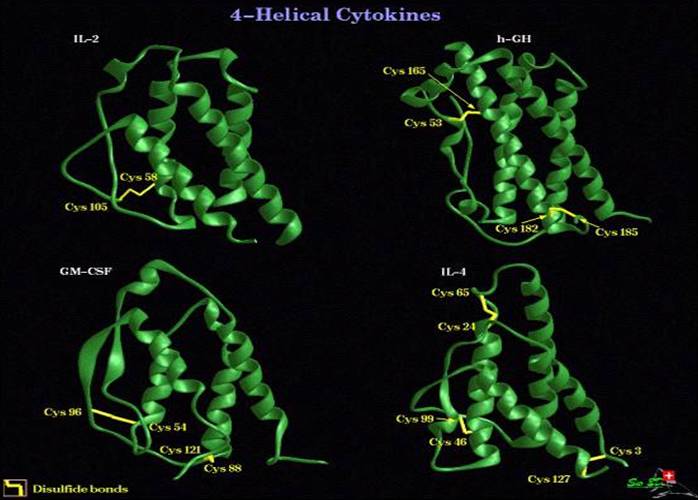
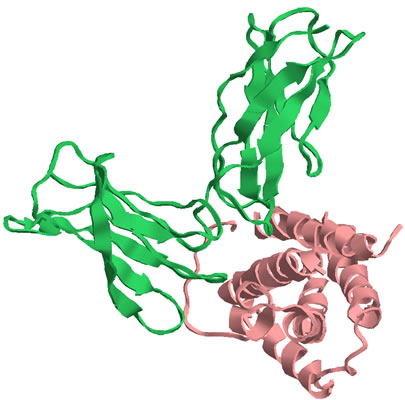
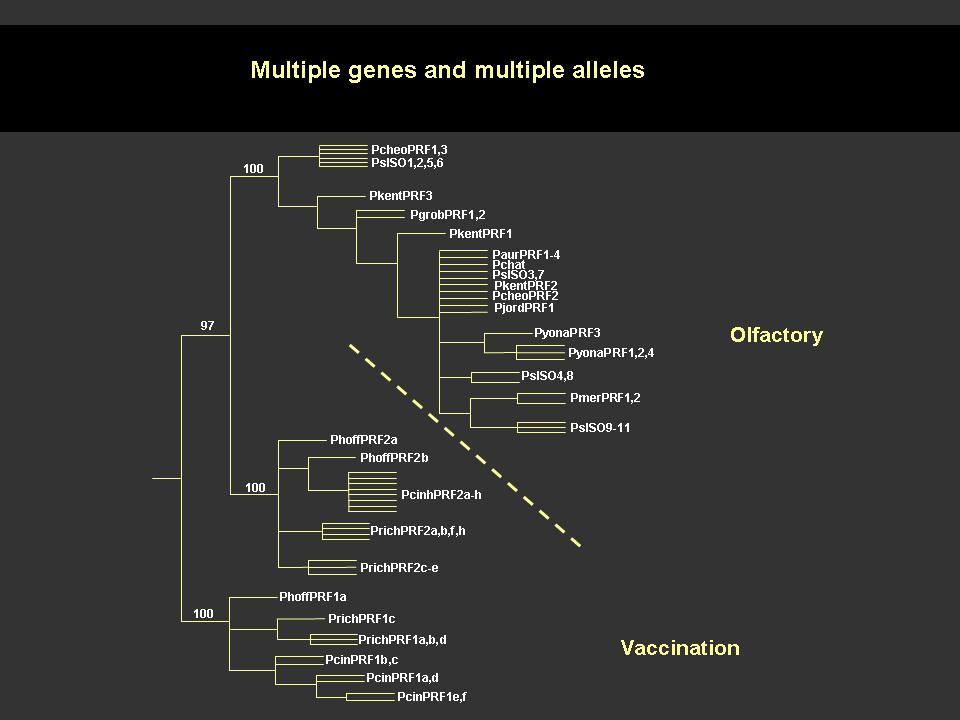
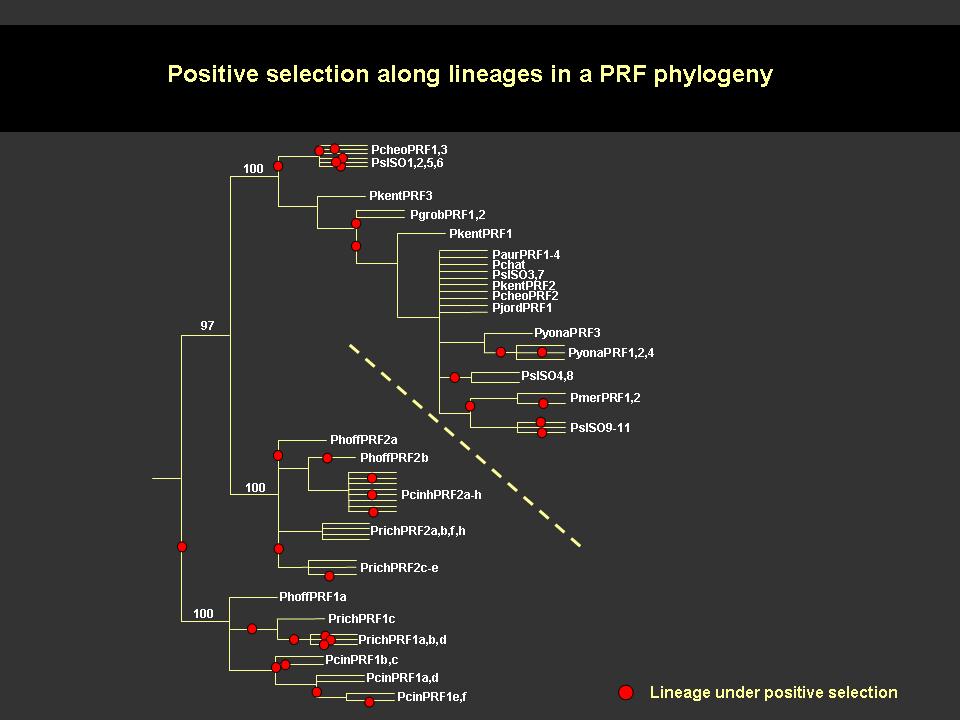
The 22kDa PRF pheromone family has been purified, characterized, sequenced, shown to bind to receptors in the vomeronasal organ (VNO) and to increase female sexual receptivity. We have developed a bacterial expression system for PRF, and recombinant PRF (rPRF) identical to one of the major natural isoforms has been shown to possess biological activity similar to the natural isoform (Houck et al. 2006b). Soluble recombinant PRF was expressed in liquid culture with remarkable efficiency (~75mg/L) and was purified using a cleavable metal-affinity tag (6xHis) followed by HPLC. Circular dichroism measurements of 3 PRF isoforms and rPRF indicate a secondary structure composition that is predominantly alpha-helix with an amount typical of the 4 alpha-helix bundle of IL-6 cytokines. The 3-dimensional structure of the rPRF will be determined. The 7 kDa PMF family contains more than 30 very negatively charged isoforms representing the products of multiple genes and allelic variants. The total amounts of various isoforms are consistent from year-to-year, but differences in the relative levels of individual isoforms suggest their expression is differentially regulated at the transcriptional and/or translational levels. We have purified and performed peptide mass fingerprinting of 16 PMF isoforms, of which 8 matched our current database of 32 unique cDNA sequences (Fig. 2). The cDNA library was prepared from mRNA obtained from the mental glands of male P. shermani during their courtship season. The 7 kDa pheromones have 8 conserved Cys residues (4 disulfide bonds; ~60 residues) with spacing similar to that for xenoxins and snake toxins, but with limited sequence homology.
Conditions for the determination of the disulfide bonding pattern were developed for a single purified isoform which resulted in the average reduction and alkylation of 1, 2 or 3 disulfide bonds. Aliquots of each were then subjected to complete digestion with trypsin, chymotrypsin or Glu-C prior to direct MALDI-TOF analysis. All of the MS data sets are consistent with a disulfide bonding pattern of 1-2, 3-6/7, 4-5, and 6/7-8, with the 3-6/7 bond being more susceptible to reduction and alkylation than the 1-2 bond. This pattern will dictate a 3-dimensional structure different from that for xenoxins and snake toxins (1-3, 2-4, 5-6, 7-8) and may represent a sub-family for courtship pheromones or a new family of 8 Cys/7 kDa proteins. MOLECULAR COEVOLUTION Comparative studies of cDNA sequences for PRF and PMF in the genus Plethodon reveal that both proteins have undergone rapid, selection-driven evolution over the last 27 million years. This continual, rapid evolution is characterized by convergent evolutionary events, gene duplication, production of pseudogenes, and the maintenance of polymorphism (Watts et al. 2004; Palmer et al. 2005, 2006). We have referred to the process producing this evolutionary pattern in male courtship pheromones as a ‘molecular tango’ and have hypothesized that it is driven by coevolution with receptors in the female VNO (Watts et al. 2004, Palmer et al. 2005). To test this coevolutionary hypothesis we prepared cDNA from female VNO tissue and used RT-PCR and 5’-RACE PCR to amplify large segments of pheromone receptors. This strategy revealed 67 distinct V2R isoforms, belonging to at least 6 families, in P. shermani. Ongoing discovery of new isoforms shows that the V2R family is as diverse in Plethodon as it is in rodents and other vertebrates. Our preliminary studies in Plethodon have also documented a type of receptor not previously reported from the VNO. These gp130 and related receptors may mediate PRF reception in the VNO because these receptors are universally utilized by those cytokines most similar to PRF (Watts et al. 2004, Bravo & Heath 2000). Both gp130 and CNTF-r from Plethodon VNO show unusually high levels of sequence diversity compared to other animals. This departure from the usual purifying selection mode for gp130 receptors and cytokines is consistent with a coevolutionary process driving diversification of PRF and these gp130 receptors. |